Help
1. About C2CDB
C2CDB collects comprehensive information about expression profile and biological mechanism of 318,158 cancer-related circRNAs from over 24 types of cancers.
Comparing with existing database, C2CDB owns several great advantages in the following aspects:
First, C2CDB quantified a total of 318,158 cancer-related circRNAs from 2,510 cancer samples of over 24 types of cancers, of which 96,597 have not been included in any other database yet. Beside 1,473 sequencing data and 591 mircroarray data from GEO database, we integrated 446 high-quality sequencing data of paired tumor and tumor-adjacent tissue from our laboratory. Meanwhile, C2CDB collects 1,181 samples of different tissues/cells and cancer cell-lines from ENCODE database. These makes C2CDB more comprehensive than current circRNA databases. In addition, the “circRNA in Cancer” section exhibits differential expression of circRNAs. In this section, users can browse differential expressed circRNAs in each dataset and obtain candidates for follow-up studies by intersection analysis of different datasets.
Second, C2CDB provides users detailed information of biological mechanism of cancer-related circRNAs, including miRNA sponge, RBP binding, coding potential, base modification, mutation, and secondary structure, covering almost all aspects of the information needed for the current research on circRNA mechanisms. Besides, visualization interface allows users to get information of circRNA mechanisms more clearly and intuitively comparing to the similar databases.
Third, unlike other databases, C2CDB focuses more on biological function and mechanism of circRNAs. C2CDB collected the experimental data from published articles, including biological function, mechanism and molecular tools like QT-qPCR primers, Northern-blot probes and shRNA sequences. To get these information accurately, all the articles about circRNAs are downloaded from NCBI Pubmed database and the experimental data in these articles were extracted by experienced circRNA researchers in our group.
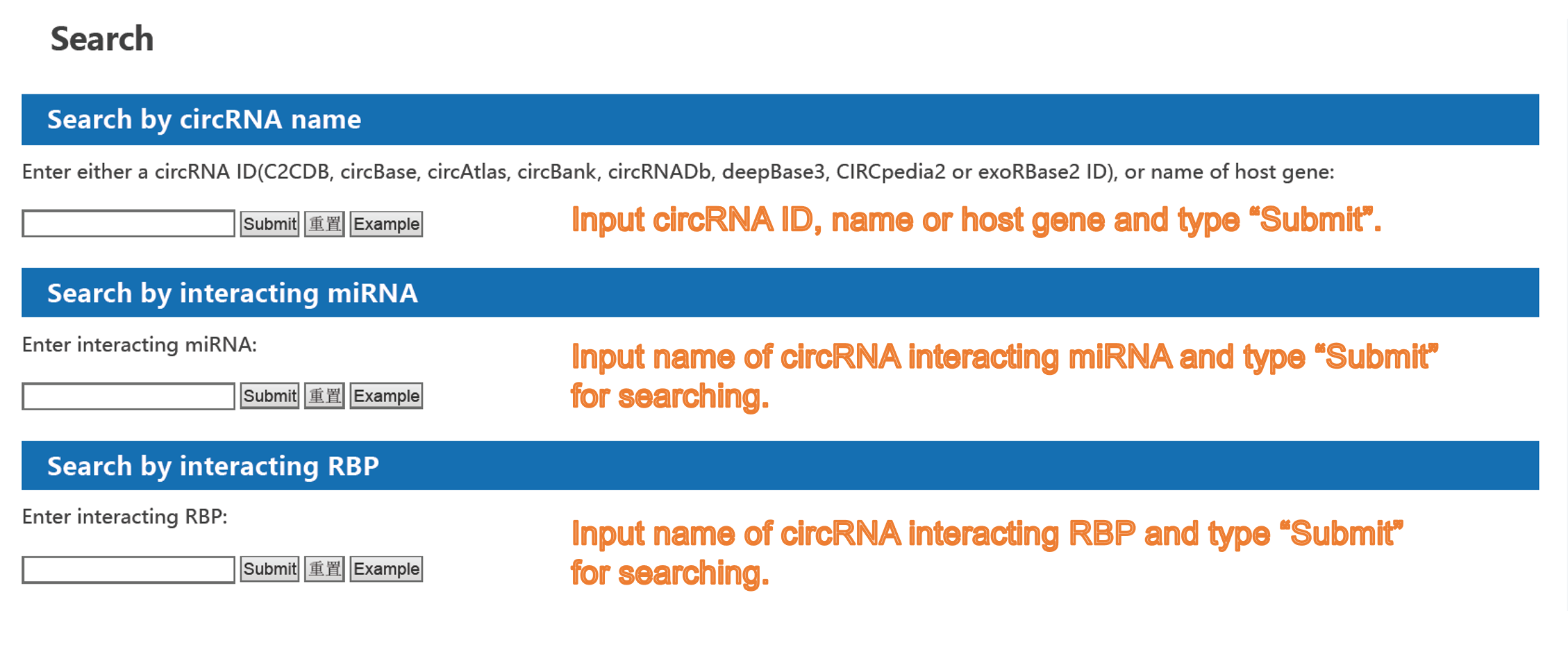
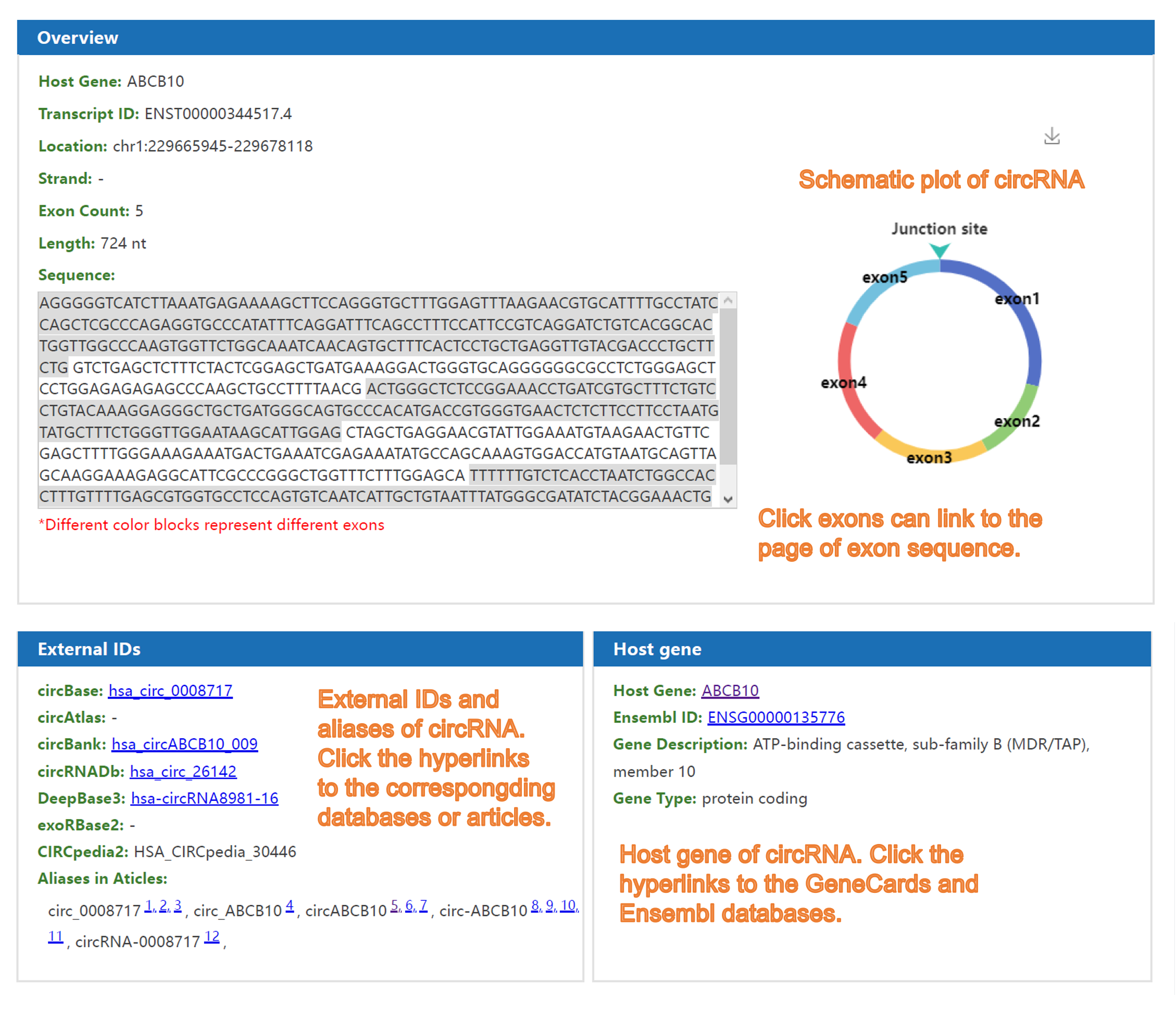
Forth, the nomenclature of a certain circRNA is not universal across databases and articles. For example, the well-known circRNA CDR1as is named as hsa_circ_0001946 in circBase, hsa_circCDR1_001 in circBank, hsa-circRNA8162 in DeepBase3, as well as circRNA7 and ciRS-7 in different articles, which makes the researchers confused. Given this, C2CDB provides users a new naming system of circRNAs which integrates the names of circRNAs from different databases and articles. Meanwhile, users can search circRNAs by name from any sources and convert names across different databases using the “ID Converter” tool. We believe it will help to solve the trouble caused by inconsistent naming methods for a long time and benefit our users.
Last but most important, to meet the needs of different users, C2CDB provides various tools for circRNA analysis. Besides the “ID Converter” mentioned above, “Genome Browser” and “BLAST” will help user searching and browsing circRNAs easier. Most notably, we build a “Integrated Analysis” tool for advanced analysis targeting current focal issues in the field of circRNAs, as circRNA-miRNA ceRNA, circRNA encoding peptides and circRNA biogenesis. It integrates mult-omics result from RNA expression correlation, CLIP-seq mapping, RNA-RNA binding prediction, RNA-protein binding prediction, and modification site recognition.
2. User guide
1) The pages and their utilities:
2) Search page of C2CDB:
3) The page of detailed information about circRNA:
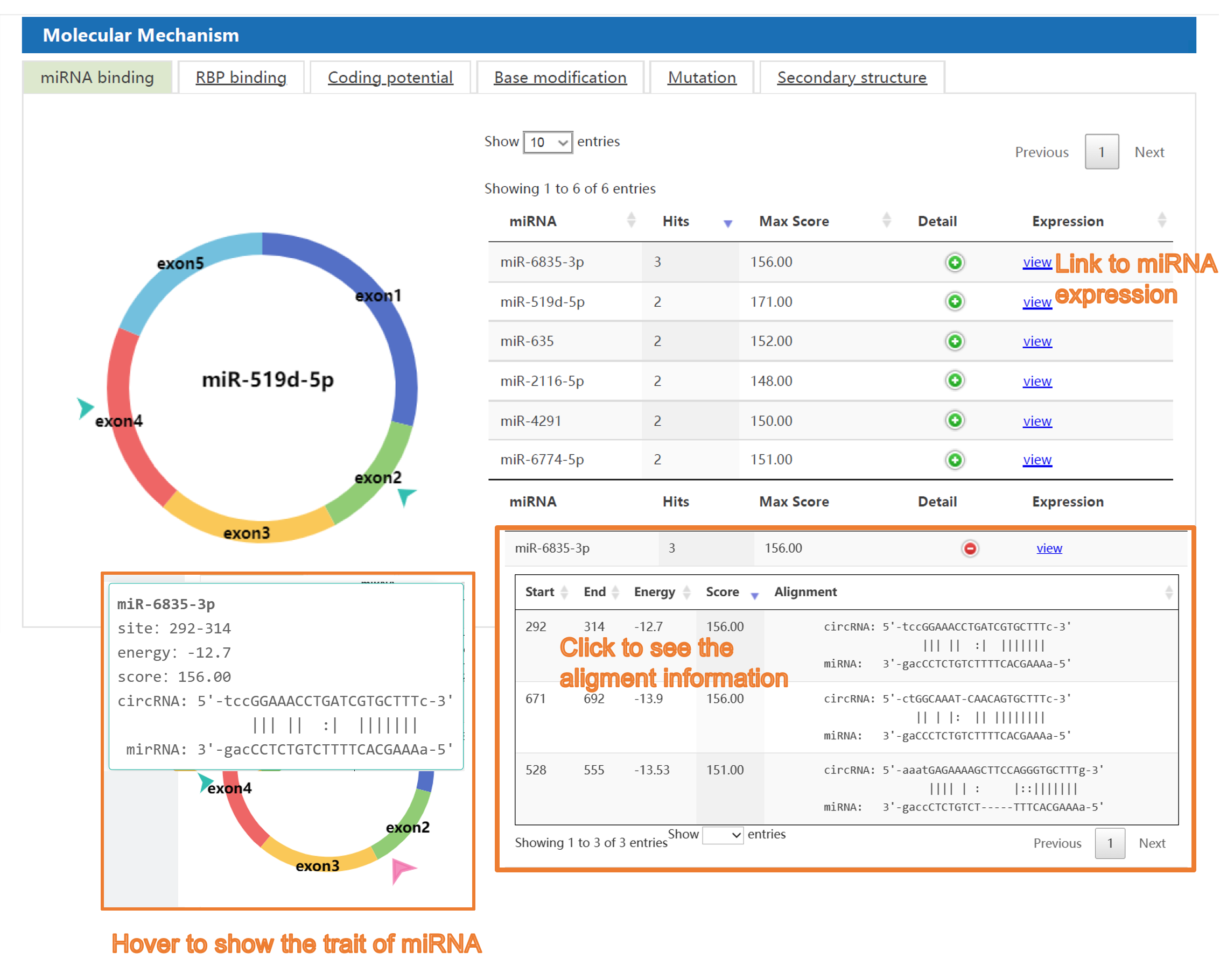
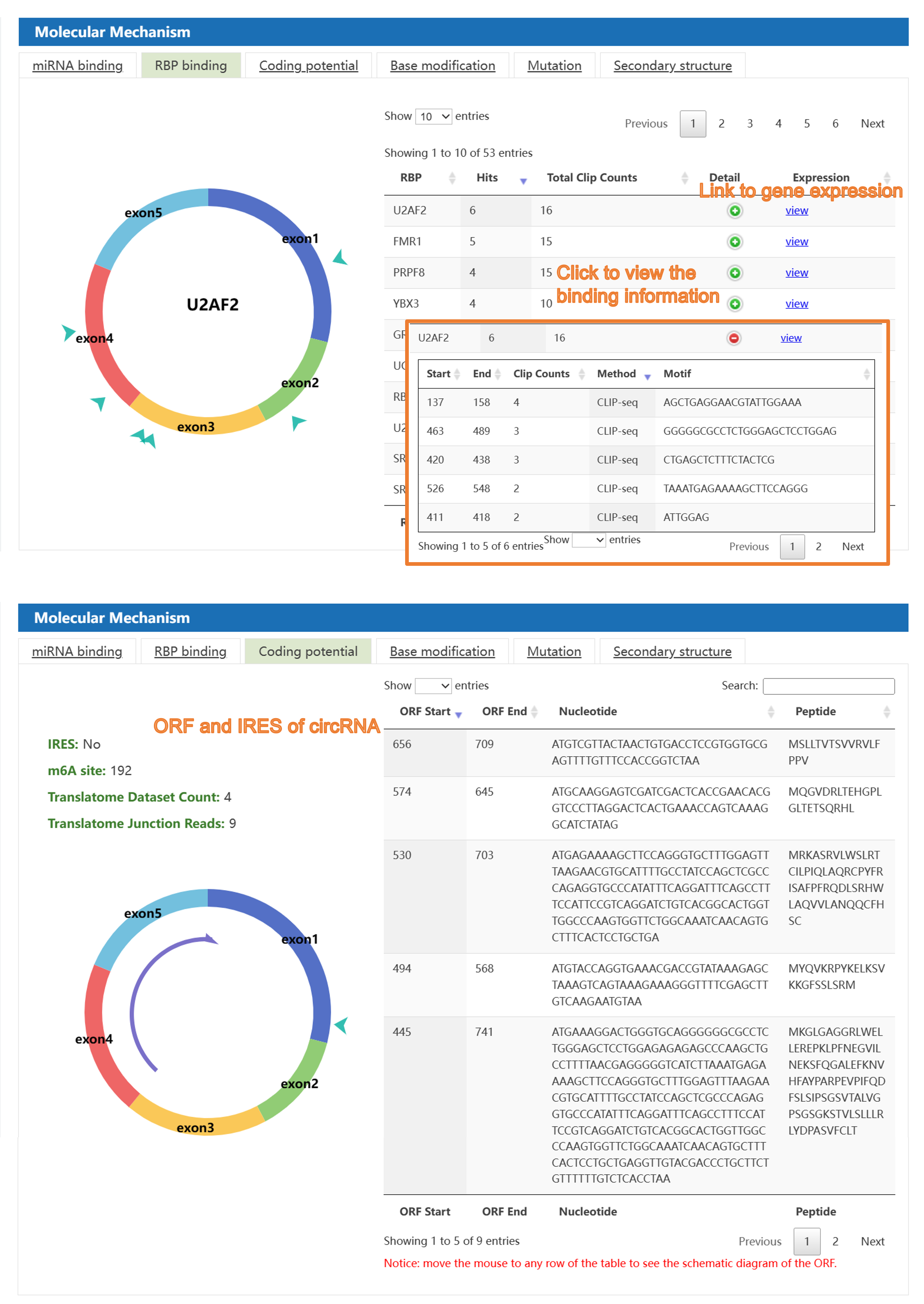
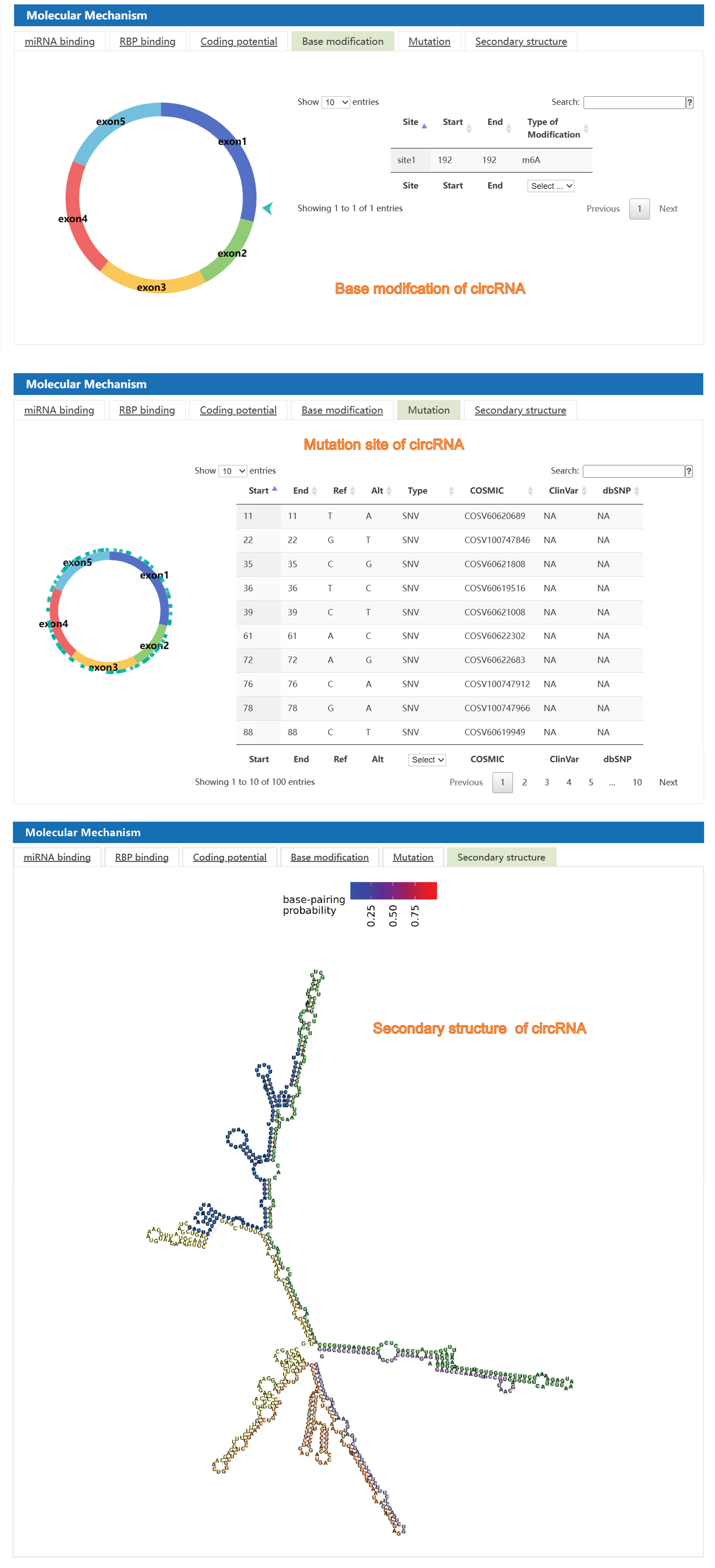
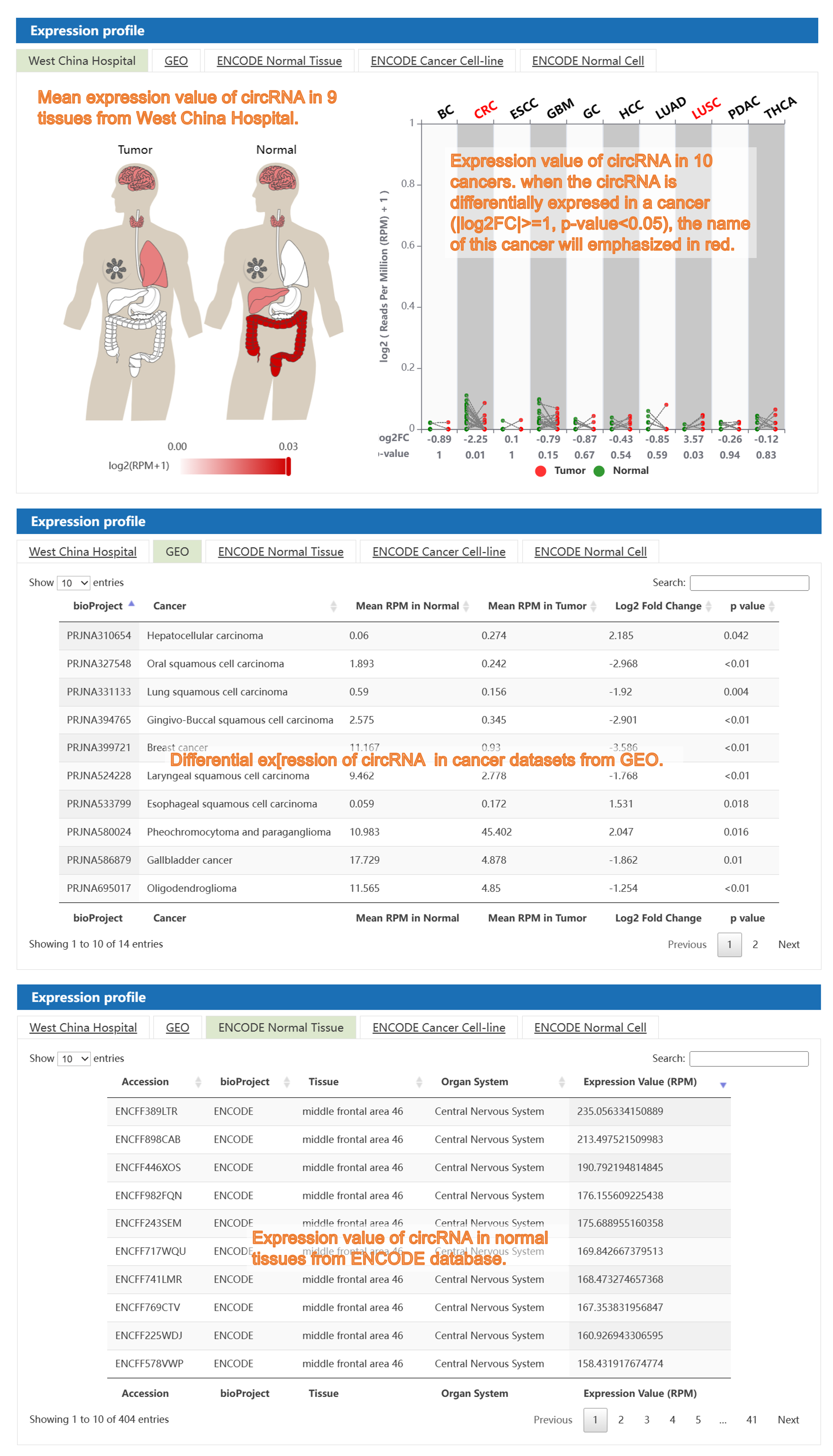
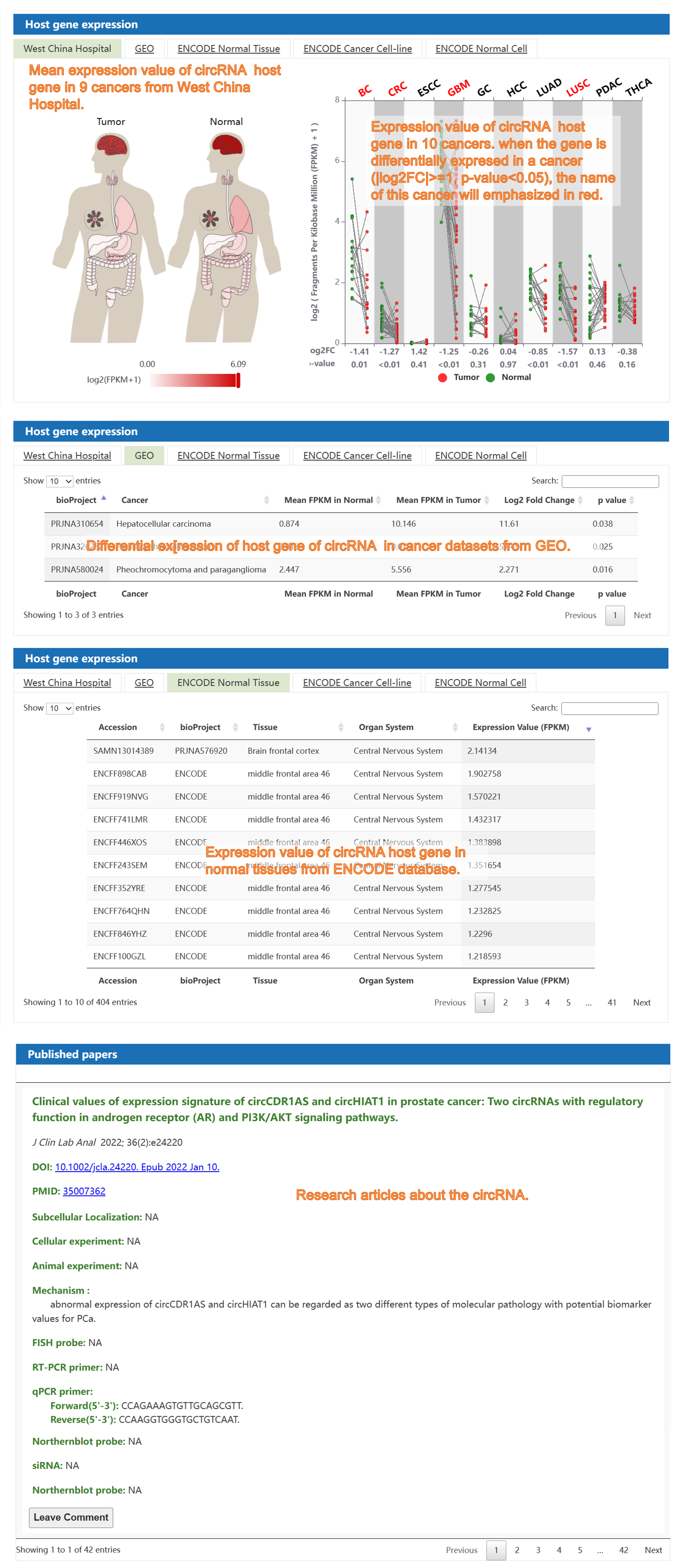
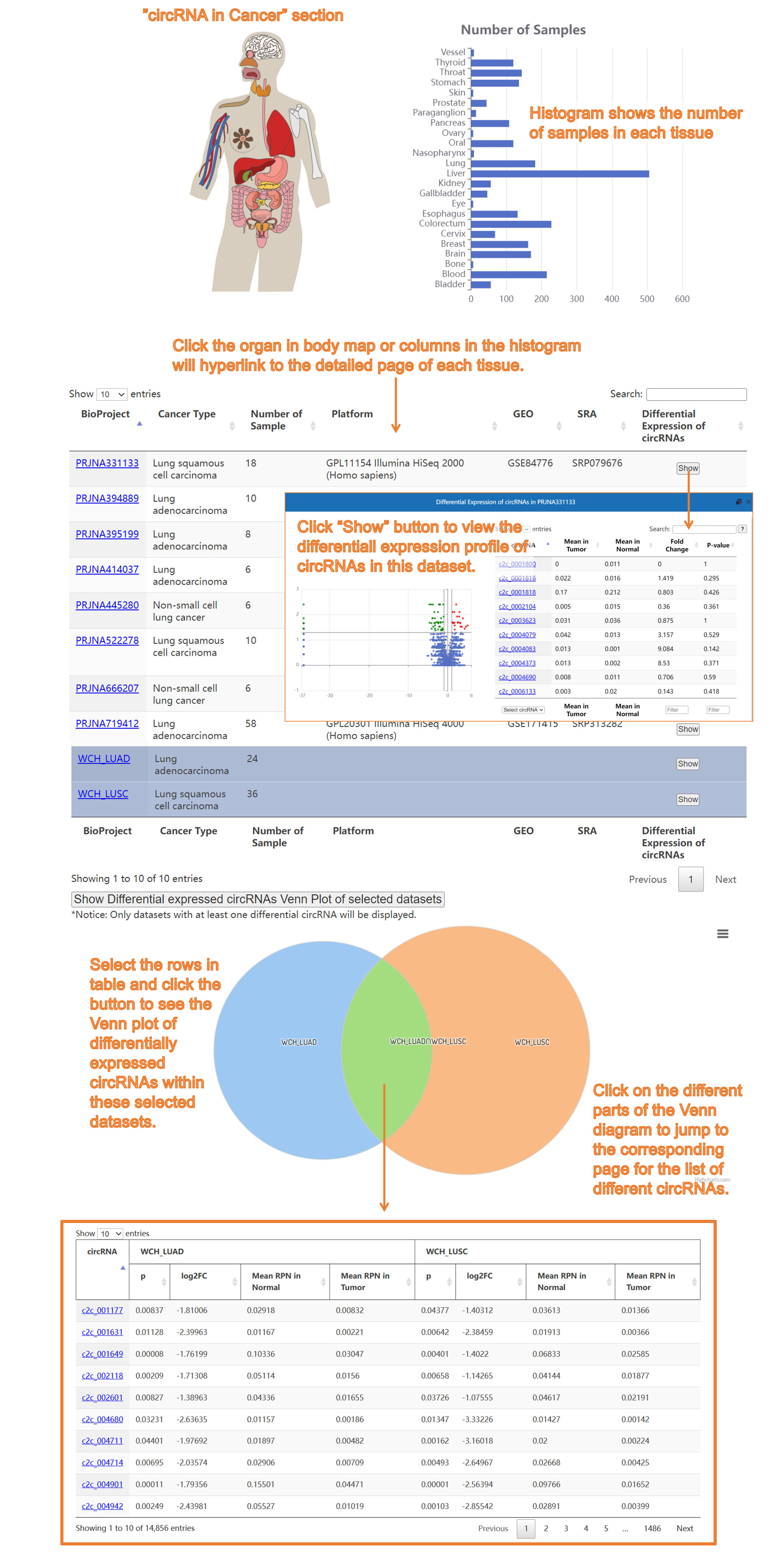
4) The "circRNA in Caner" section:
5) ID Converter:

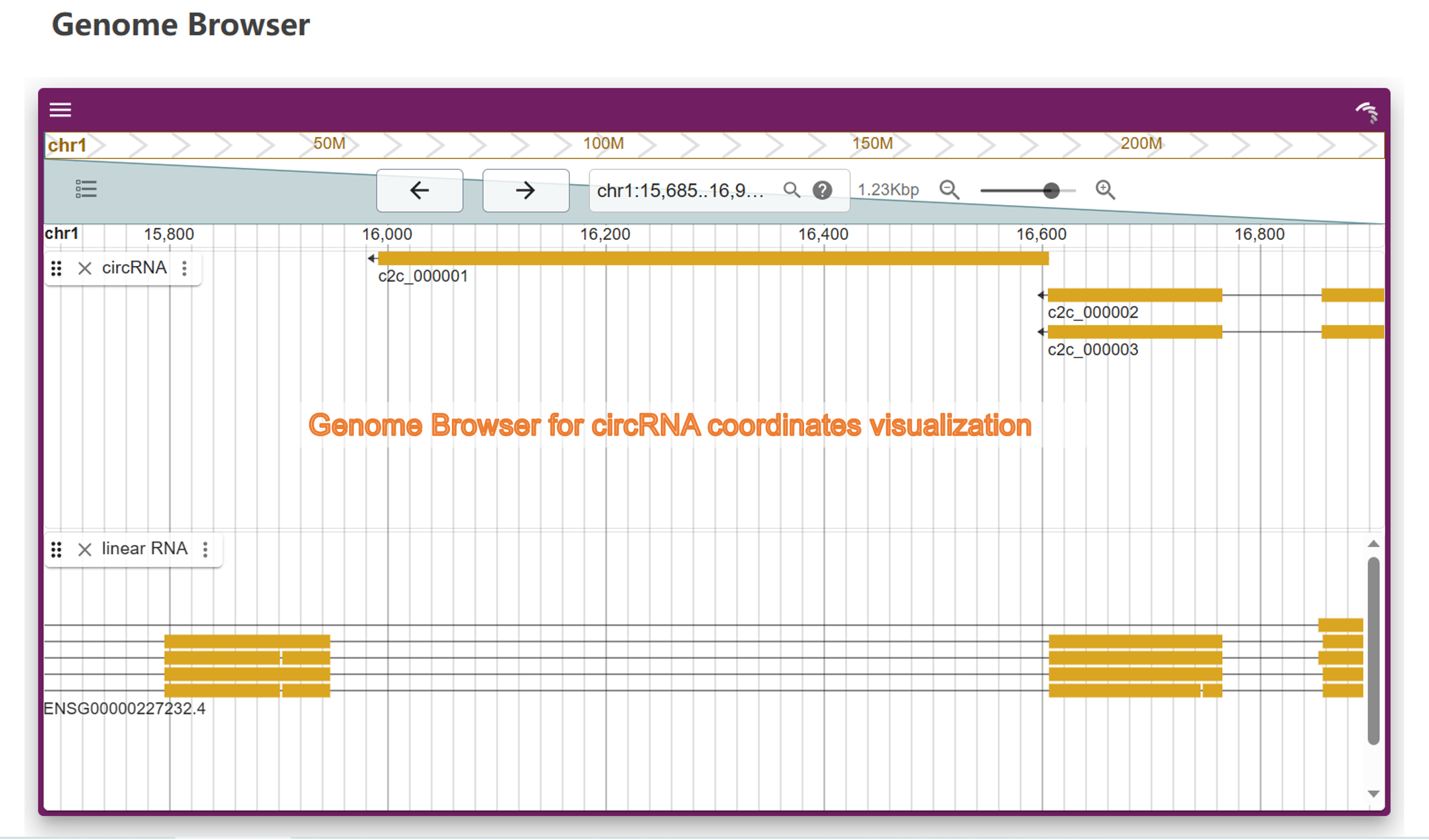
6) Genome Browser:
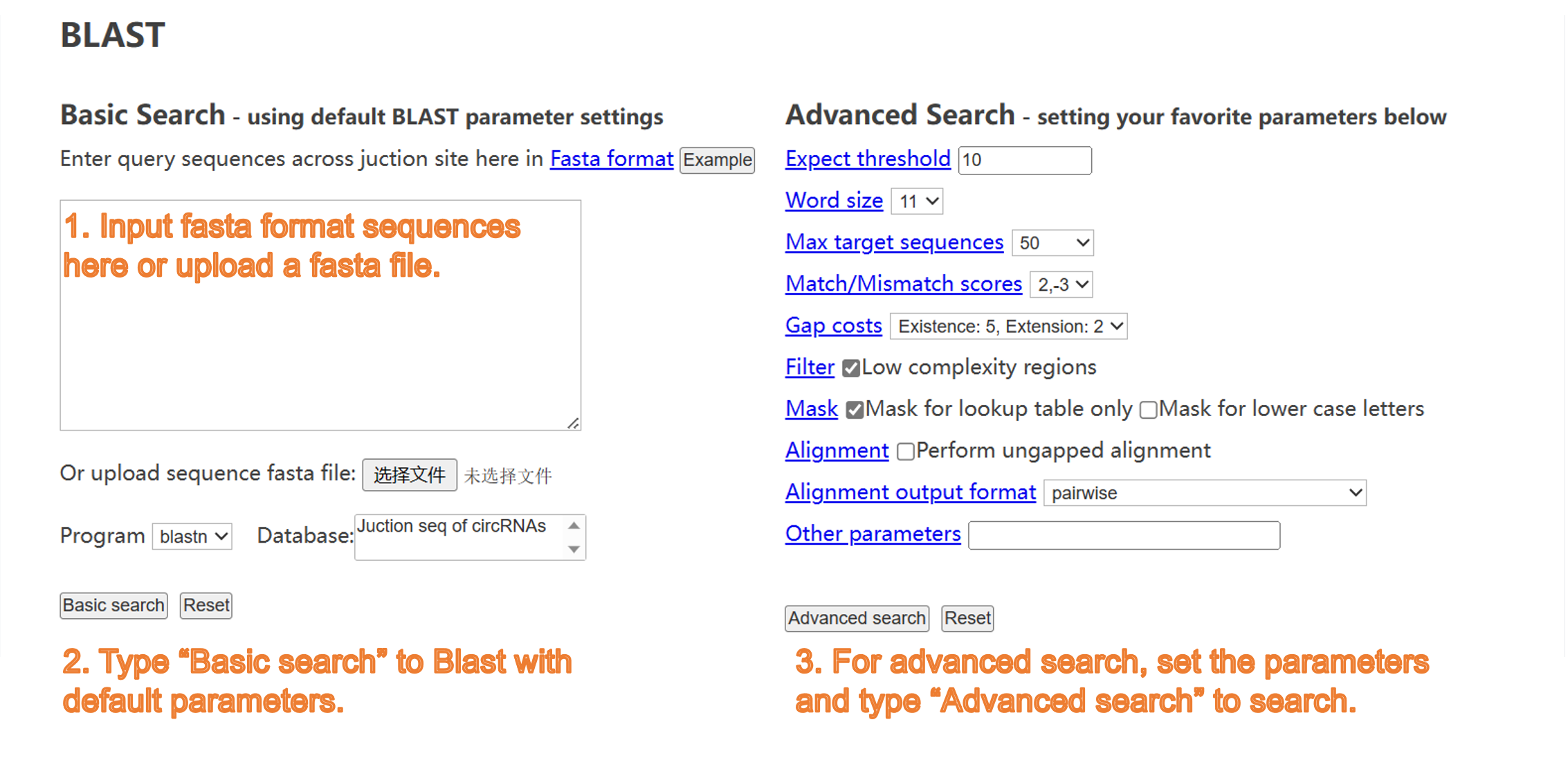
7) BLAST tool:
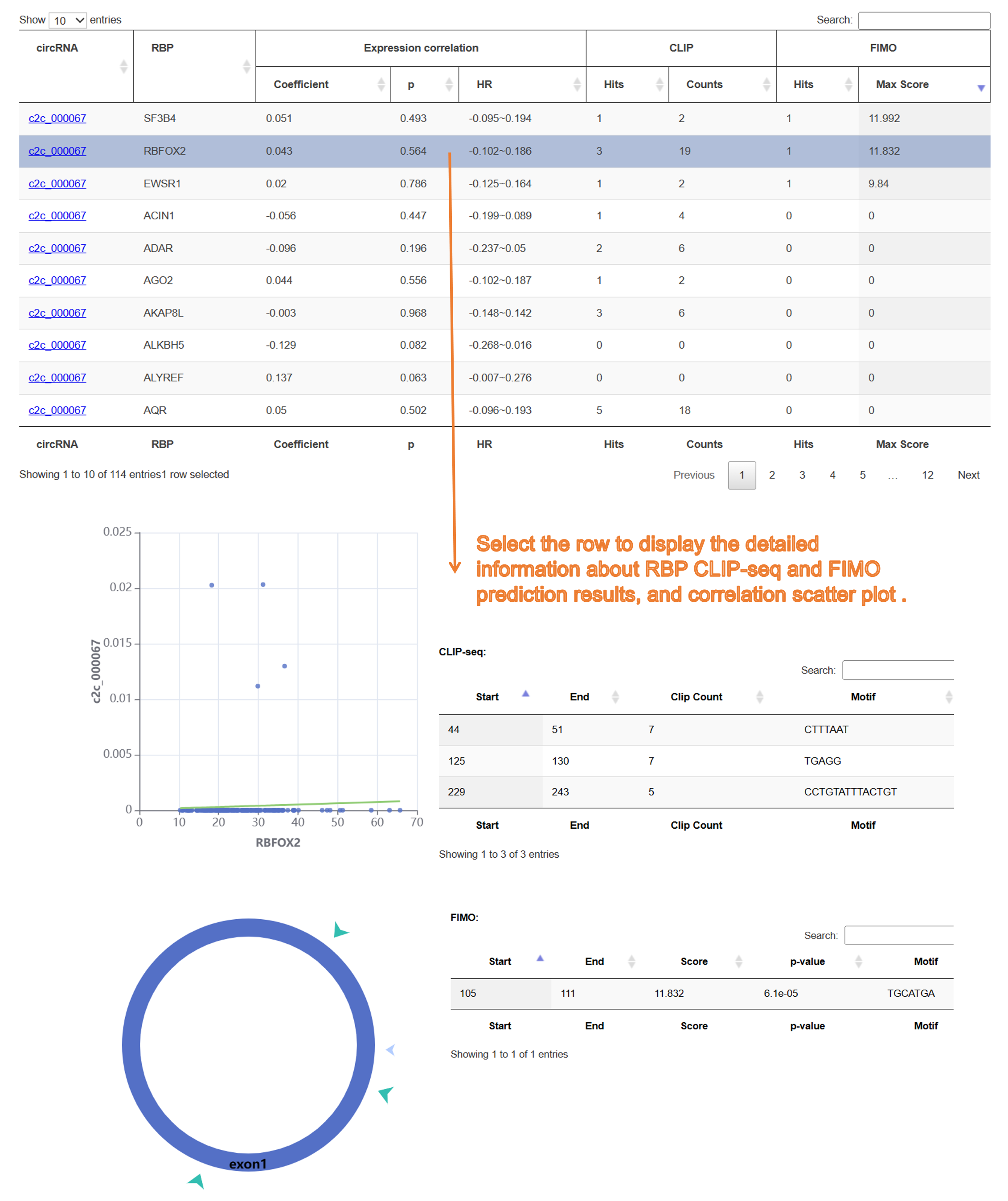
8) Integrated Analysis tool:

9) Result page of "circRNA-miRNA ceRNA":
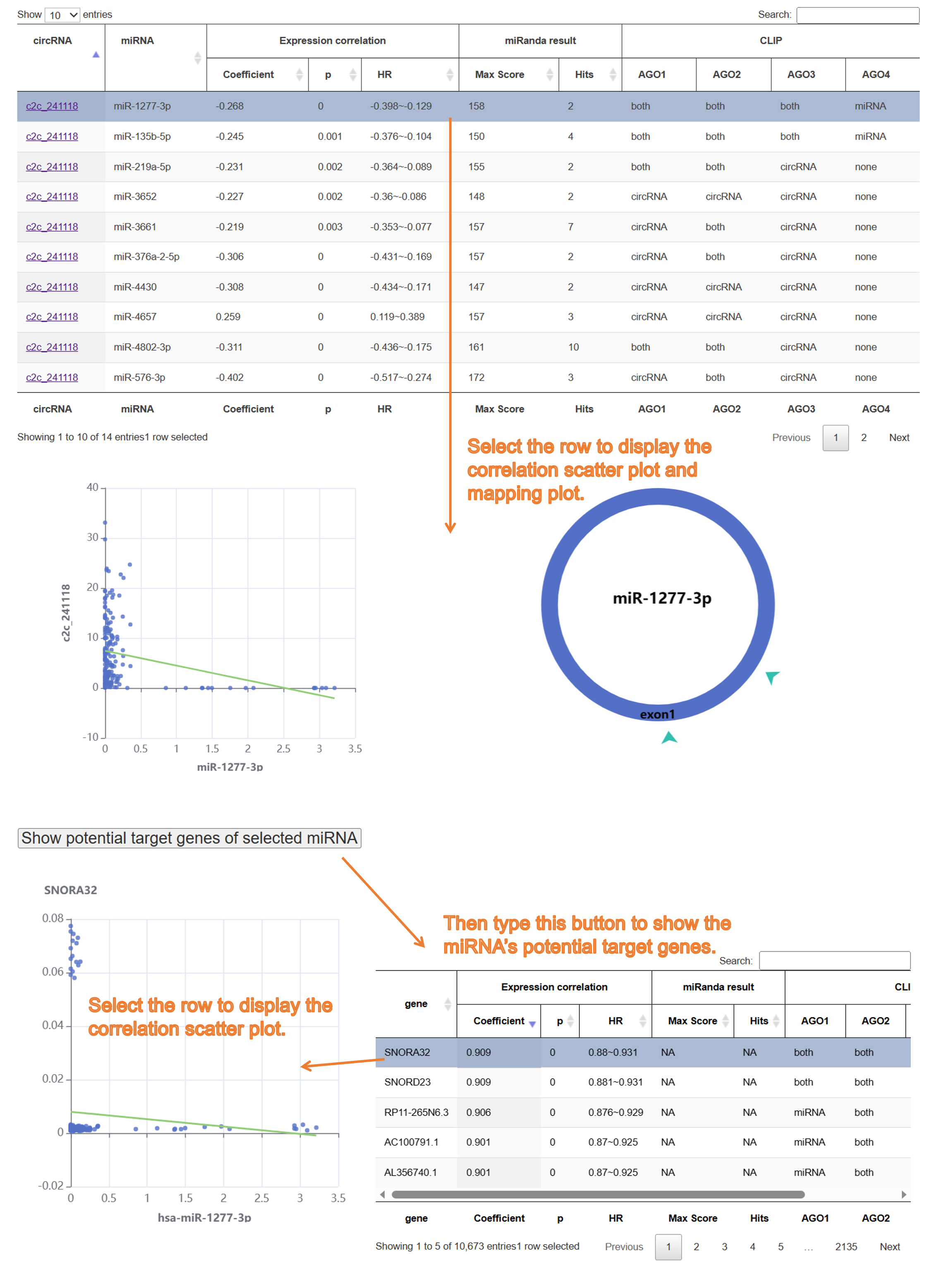
10) Result page of "circRNA encoding peptides":
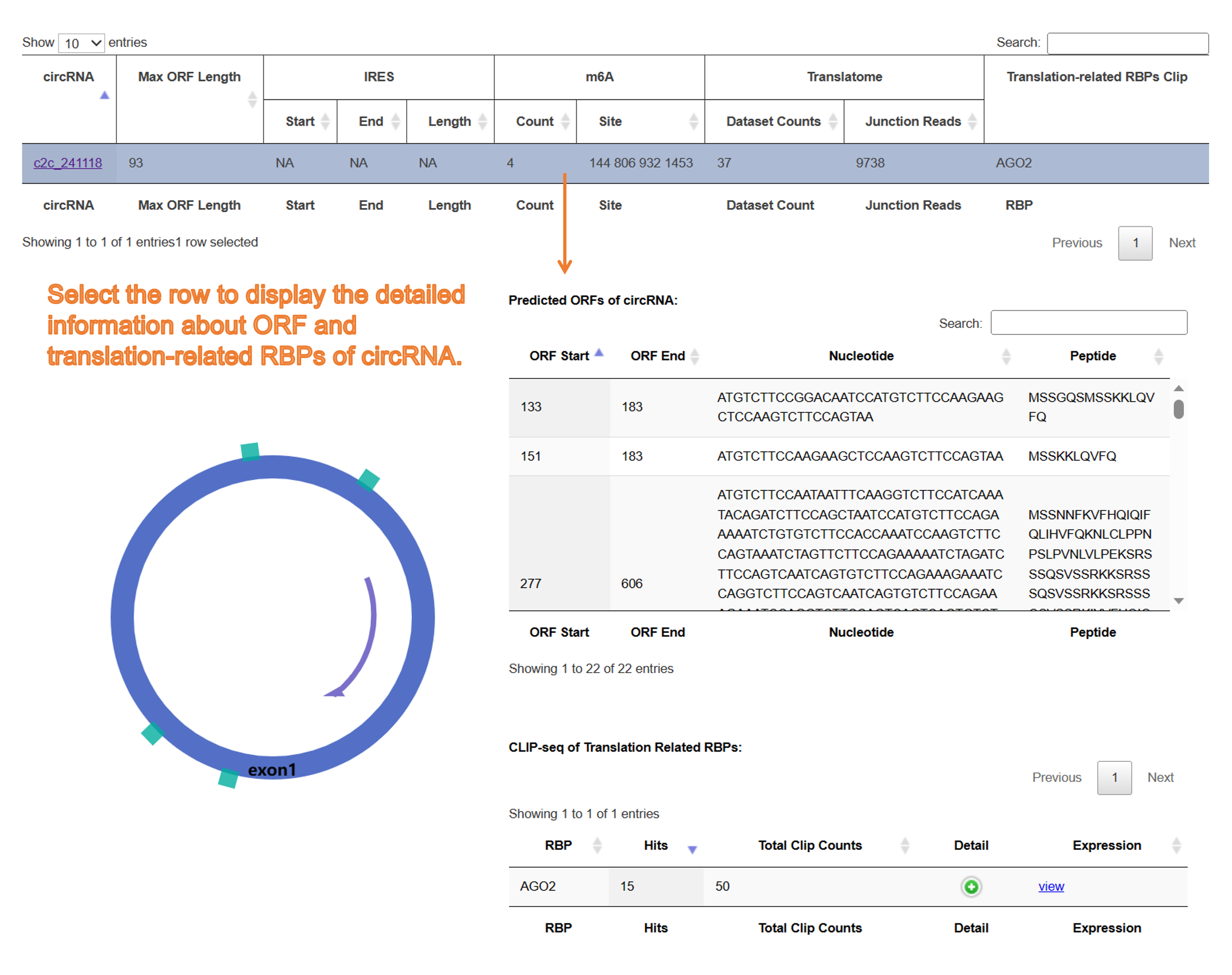
11) Result page of "circRNA biogenesis":